Novel method for creating sequence-defined polymers

RESEARCHERS at Imperial College London, UK have developed a new class of multifunctional sequence-defined polymers. The polymers were created using a novel, universal method of synthesis which enabled full control over the final polymer sequence.
Biopolymers, such as DNA and proteins, occur naturally in the body and are composed of thousands of molecules or more. In nature they are created with a high level of precision and provide structural and functional diversity. Chemists have developed synthetic polymers that work by mimicking natural sequences to deliver additional functions. Polyethers are favoured for use in medicine and surgery because of their excellent water solubility and biocompatibility.
Synthetic polymers are typically made with one type of monomer, but sometimes more than one type is used. When more than one type of monomer is involved, traditional synthesis methods are unable to achieve the sequence precision observed in nature.
Chemical engineers and chemists in the Livingston Group at Imperial used a novel process called “Nanostar Sieving” to create polymers with “extraordinary” precision. The process combines liquid-phase synthesis with molecular sieving and enables researchers to have complete control over the monomer sequence in the resulting polymer.
The researchers used four chiral pentaethylene glycols with diverse side groups as the building block for polyethylene glycol (PEG) production. The building blocks were assigned the coding letters N, O, L, and D, respectively.
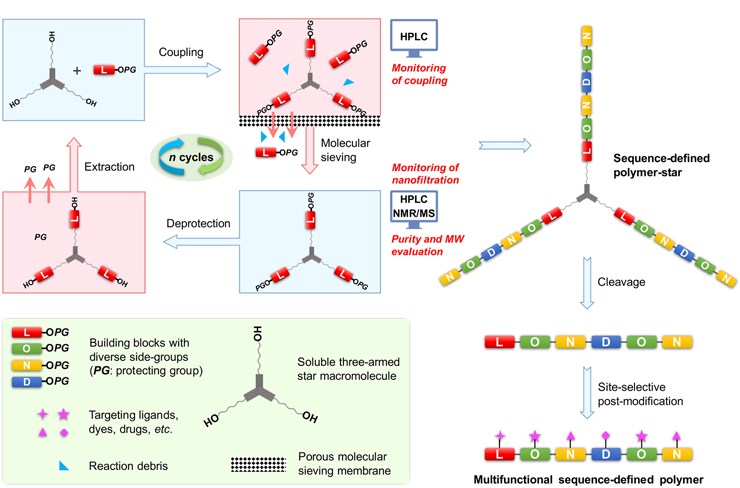
To create the PEG molecules a soluble three-armed star macromolecule, referred to as the hub, was coupled with building blocks with protecting groups at one end. The protecting groups on the monomers were used to prevent uncontrolled chain extension. Coupling was followed by removal of excess monomers and other reaction debris by molecular sieving. The protecting groups were then removed and extracted. Further coupling could then be carried out to achieve chain extension.
By this method the researchers created sequence-defined polyether stars with the monomer sequences LONDON and LOLLOL using a chain extension cycle.
The method also allows for site-selective post-synthetic modification. It provides a versatile pathway by which to control side-chain structures and functions.
The researchers expect that this platform will enable the creation of novel sequence-defined polymers which could be used in real-life applications. It opens up the potential uses for polyethers in a range of applications such as healthcare, nanotechnology, and information storage.
Ruijiao Dong, associate researcher in the department of chemical engineering at Imperial, told a university reporter: “It’s an exciting piece of research because we’re one step closer to creating a well-defined hierarchical structure which mimics that found in nature. By developing a universal strategy for precisely controlling sequence in polymers we’re in theory able to do many things, as the sequence plays a crucial role in information storage and function expression, offering potential for a wide range of applications. We are excited to test the limits of its functionality with further research, and to further develop and automate the Nanostar Sieving.”
The team will now explore the practical application of the new polyether, and experiment with others. They aim to create a biodegradable version, control the three-dimensional structure of the final polymers, and develop methods to read/write for information storage and polymer synthesis.
Nature Chemistry: http://doi.org/cx5w
Recent Editions
Catch up on the latest news, views and jobs from The Chemical Engineer. Below are the four latest issues. View a wider selection of the archive from within the Magazine section of this site.




